Usage¶
Because PRIMAP2 builds on xarray, all xarray functionality is available right away. Additional functionality is provided in the primap2 package and in the pr namespace on xarray objects. In this section, we will present examples of PRIMAP2 usage.
Importing¶
[1]:
import primap2 # injects the "pr" namespace into xarray
[2]:
# set up logging for the docs - don't show debug messages
import sys
from loguru import logger
logger.remove()
logger.add(sys.stderr, level="INFO")
[2]:
1
Loading Datafiles¶
Loading from netcdf files¶
The native storage format of PRIMAP2 are netcdf5 files, and datasets can be written to and loaded from netcdf5 files using PRIMAP2 functions. We will load the “minimal” and “opulent” Datasets from the data format section:
[3]:
ds_min = primap2.open_dataset("minimal_ds.nc")
ds = primap2.open_dataset("opulent_ds.nc")
ds
[3]:
<xarray.Dataset>
Dimensions: (time: 21, area (ISO3): 4, category (IPCC 2006): 8,
animal (FAOSTAT): 3, product (FAOSTAT): 2,
scenario (FAOSTAT): 2, provenance: 1, model: 1,
source: 2)
Coordinates:
* animal (FAOSTAT) (animal (FAOSTAT)) <U5 'cow' 'swine' 'goat'
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* category (IPCC 2006) (category (IPCC 2006)) <U3 '0' '1' '2' ... '1.A' '1.B'
category_names (category (IPCC 2006)) <U14 'total' ... 'light indu...
* model (model) <U8 'FANCYFAO'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* provenance (provenance) <U9 'projected'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* source (source) <U8 'RAND2020' 'RAND2021'
* time (time) datetime64[ns] 2000-01-01 ... 2020-01-01
Data variables:
CH4 (time, area (ISO3), category (IPCC 2006), animal (FAOSTAT), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CH4·Gg/a] ...
CO2 (time, area (ISO3), category (IPCC 2006), animal (FAOSTAT), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CO2·Gg/a] ...
SF6 (time, area (ISO3), category (IPCC 2006), animal (FAOSTAT), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [Gg·SF6/a] ...
SF6 (SARGWP100) (time, area (ISO3), category (IPCC 2006), animal (FAOSTAT), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CO2·Gg/a] ...
population (time, area (ISO3), provenance, model, source) float64 [] ...
Attributes:
area: area (ISO3)
cat: category (IPCC 2006)
comment: GHG inventory data ...
contact: lol_no_one_will_answer@example.com
entity_terminology: primap2
history: 2021-01-14 14:50 data invented\n2021-01-14 14:51 add...
institution: PIK
references: doi:10.1012
rights: Use however you want.
scen: scenario (FAOSTAT)
sec_cats: ['animal (FAOSTAT)', 'product (FAOSTAT)']
title: Completely invented GHG inventory dataAccessing metadata¶
Metadata is stored in the attrs of Datasets, and you can of course access it directly there. Additionally, you can access the PRIMAP2 metadata directly under the .pr namespace, which has the advantage that autocompletion works in ipython and IDEs and typos will be caught immediately.
[4]:
ds.pr.title
[4]:
'Completely invented GHG inventory data'
[5]:
ds.pr.title = "Another title"
ds.pr.title
[5]:
'Another title'
Selecting data¶
Data can be selected using the xarray indexing methods, but PRIMAP2 also provides own versions of some of xarray’s selection methods which work using the dimension names without the category set.
Getitem¶
The following selections both select the same:
[6]:
ds["area (ISO3)"]
[6]:
<xarray.DataArray 'area (ISO3)' (area (ISO3): 4)> array(['COL', 'ARG', 'MEX', 'BOL'], dtype='<U3') Coordinates: * area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
[7]:
ds.pr["area"]
[7]:
<xarray.DataArray 'area (ISO3)' (area (ISO3): 4)> array(['COL', 'ARG', 'MEX', 'BOL'], dtype='<U3') Coordinates: * area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
The loc Indexer¶
Similarly, a version of the loc indexer is provided which works with the bare dimension names:
[8]:
ds.pr.loc[{"time": slice("2002", "2005"), "animal": "cow"}]
[8]:
<xarray.Dataset>
Dimensions: (time: 4, area (ISO3): 4, category (IPCC 2006): 8,
product (FAOSTAT): 2, scenario (FAOSTAT): 2,
provenance: 1, model: 1, source: 2)
Coordinates:
animal (FAOSTAT) <U5 'cow'
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* category (IPCC 2006) (category (IPCC 2006)) <U3 '0' '1' '2' ... '1.A' '1.B'
category_names (category (IPCC 2006)) <U14 'total' ... 'light indu...
* model (model) <U8 'FANCYFAO'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* provenance (provenance) <U9 'projected'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* source (source) <U8 'RAND2020' 'RAND2021'
* time (time) datetime64[ns] 2002-01-01 ... 2005-01-01
Data variables:
CH4 (time, area (ISO3), category (IPCC 2006), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CH4·Gg/a] ...
CO2 (time, area (ISO3), category (IPCC 2006), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CO2·Gg/a] ...
SF6 (time, area (ISO3), category (IPCC 2006), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [Gg·SF6/a] ...
SF6 (SARGWP100) (time, area (ISO3), category (IPCC 2006), product (FAOSTAT), scenario (FAOSTAT), provenance, model, source) float64 [CO2·Gg/a] ...
population (time, area (ISO3), provenance, model, source) float64 [] ...
Attributes:
area: area (ISO3)
cat: category (IPCC 2006)
comment: GHG inventory data ...
contact: lol_no_one_will_answer@example.com
entity_terminology: primap2
history: 2021-01-14 14:50 data invented\n2021-01-14 14:51 add...
institution: PIK
references: doi:10.1012
rights: Use however you want.
scen: scenario (FAOSTAT)
sec_cats: ['animal (FAOSTAT)', 'product (FAOSTAT)']
title: Another titleIt also works on DataArrays:
[9]:
da = ds["CO2"]
da.pr.loc[
{
"time": slice("2002", "2005"),
"animal": "cow",
"category": "0",
"area": "COL",
}
]
[9]:
<xarray.DataArray 'CO2' (time: 4, product (FAOSTAT): 2, scenario (FAOSTAT): 2,
provenance: 1, model: 1, source: 2)>
<Quantity([[[[[[0.13213244 0.30232155]]]
[[[0.56331297 0.86483736]]]]
[[[[0.4988332 0.31410627]]]
[[[0.10251511 0.81517807]]]]]
[[[[[0.17221409 0.44064751]]]
[[[0.08172373 0.93020094]]]]
...
[[[[0.41901311 0.35808953]]]
[[[0.66955278 0.09849324]]]]]
[[[[[0.54763585 0.6474731 ]]]
[[[0.89670035 0.70107409]]]]
[[[[0.78242851 0.08874086]]]
[[[0.70000787 0.16270348]]]]]], 'CO2 * gigagram / year')>
Coordinates:
animal (FAOSTAT) <U5 'cow'
area (ISO3) <U3 'COL'
category (IPCC 2006) <U3 '0'
category_names <U14 'total'
* model (model) <U8 'FANCYFAO'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* provenance (provenance) <U9 'projected'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* source (source) <U8 'RAND2020' 'RAND2021'
* time (time) datetime64[ns] 2002-01-01 ... 2005-01-01
Attributes:
entity: CO2Setting data¶
PRIMAP2 provides a unified API to introduce new data values, fill missing information, and overwrite existing information: the da.pr.set function and its sibling ds.pr.set.
The basic signature of the set functions is set(dimension, keys, values), and it returns the changed object without changing the original one. Use it like this:
[10]:
da = ds_min["CO2"].loc[{"time": slice("2000", "2005")}]
da
[10]:
<xarray.DataArray 'CO2' (time: 6, area (ISO3): 4, source: 1)>
<Quantity([[[0.66352018]
[0.91683183]
[0.10638745]
[0.61416341]]
[[0.33822533]
[0.68233863]
[0.73953851]
[0.79057726]]
[[0.11409101]
[0.25072807]
[0.85044897]
[0.40873907]]
[[0.80147707]
[0.85209597]
[0.71521289]
[0.08031265]]
[[0.61018888]
[0.88112521]
[0.55500009]
[0.11105145]]
[[0.81245612]
[0.14774848]
[0.14072429]
[0.13586946]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2005-01-01
Attributes:
entity: CO2[11]:
import numpy as np
from primap2 import ureg
modified = da.pr.set(
"area", "CUB", np.linspace(0, 20, 6) * ureg("Gg CO2 / year")
)
modified
[11]:
<xarray.DataArray 'CO2' (time: 6, area (ISO3): 5, source: 1)>
<Quantity([[[ 0.91683183]
[ 0.61416341]
[ 0.66352018]
[ 0. ]
[ 0.10638745]]
[[ 0.68233863]
[ 0.79057726]
[ 0.33822533]
[ 4. ]
[ 0.73953851]]
[[ 0.25072807]
[ 0.40873907]
[ 0.11409101]
[ 8. ]
[ 0.85044897]]
[[ 0.85209597]
[ 0.08031265]
[ 0.80147707]
[12. ]
[ 0.71521289]]
[[ 0.88112521]
[ 0.11105145]
[ 0.61018888]
[16. ]
[ 0.55500009]]
[[ 0.14774848]
[ 0.13586946]
[ 0.81245612]
[20. ]
[ 0.14072429]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'ARG' 'BOL' 'COL' 'CUB' 'MEX'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2005-01-01
Attributes:
entity: CO2By default, existing non-NaN values are not overwritten:
[12]:
try:
da.pr.set("area", "COL", np.linspace(0, 20, 6) * ureg("Gg CO2 / year"))
except ValueError as err:
print(err)
Values {'COL'} for 'area (ISO3)' already exist and contain data. Use existing='overwrite' or 'fillna' to avoid this error.
You can overwrite existing values by specifying existing="overwrite" to overwrite all values or existing="fillna" to overwrite only NaNs.
[13]:
da.pr.set(
"area",
"COL",
np.linspace(0, 20, 6) * ureg("Gg CO2 / year"),
existing="overwrite",
)
[13]:
<xarray.DataArray 'CO2' (time: 6, area (ISO3): 4, source: 1)>
<Quantity([[[ 0.91683183]
[ 0.61416341]
[ 0. ]
[ 0.10638745]]
[[ 0.68233863]
[ 0.79057726]
[ 4. ]
[ 0.73953851]]
[[ 0.25072807]
[ 0.40873907]
[ 8. ]
[ 0.85044897]]
[[ 0.85209597]
[ 0.08031265]
[12. ]
[ 0.71521289]]
[[ 0.88112521]
[ 0.11105145]
[16. ]
[ 0.55500009]]
[[ 0.14774848]
[ 0.13586946]
[20. ]
[ 0.14072429]]], 'CO2 * gigagram / year')>
Coordinates:
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2005-01-01
* area (ISO3) (area (ISO3)) <U3 'ARG' 'BOL' 'COL' 'MEX'
* source (source) <U8 'RAND2020'
Attributes:
entity: CO2By default, the set() function extends the specified dimension automatically to accommodate new values if not all key values are in the specified dimension yet. You can change this by specifying new="error", which will raise a KeyError if any of the keys is not found:
[14]:
try:
da.pr.set(
"area",
["COL", "CUB"],
np.linspace(0, 20, 6) * ureg("Gg CO2 / year"),
existing="overwrite",
new="error",
)
except KeyError as err:
print(err)
"Values {'CUB'} not in 'area (ISO3)', use new='extend' to automatically insert new values into dim."
In particular, the set() functions can also be used with xarray’s arithmetic functions to derive values from existing data and store the result in the Dataset. As an example, we will derive better values for category 0 by adding all its subcategories and store the result.
First, let’s see the current data for a small subset of the data:
[15]:
sel = {
"area": "COL",
"category": ["0", "1", "2", "3", "4", "5"],
"animal": "cow",
"product": "milk",
"scenario": "highpop",
"source": "RAND2020",
}
subset = ds.pr.loc[sel].squeeze()
# TODO: currently, plotting with units still emits a warning
import warnings
with warnings.catch_warnings():
warnings.simplefilter("ignore")
subset["CO2"].plot.line(x="time", hue="category (IPCC 2006)")
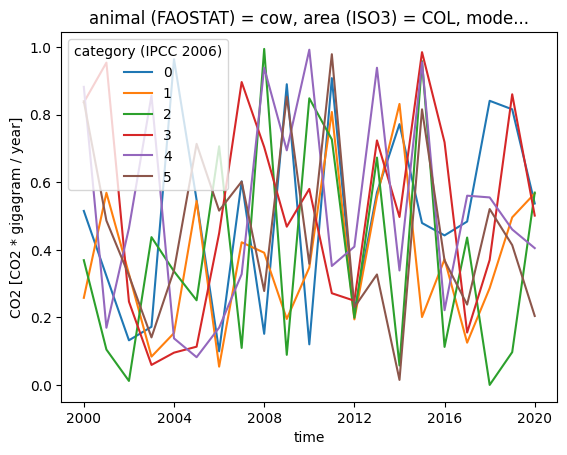
While it is hard to see any details in this plot, it is clearly visible that category 0 is not the sum of the other categories (which should not come as a surprise since we generated the data at random).
We will now recompute category 0 for the entire dataset using set():
[16]:
cat0_new = ds.pr.loc[{"category": ["1", "2", "3", "4", "5"]}].pr.sum(
"category"
)
ds = ds.pr.set(
"category",
"0",
cat0_new,
existing="overwrite",
)
# plot a small subset of the result
subset = ds.pr.loc[sel].squeeze()
# TODO: currently, plotting with units still emits a warning
import warnings
with warnings.catch_warnings():
warnings.simplefilter("ignore")
subset["CO2"].plot.line(x="time", hue="category (IPCC 2006)")
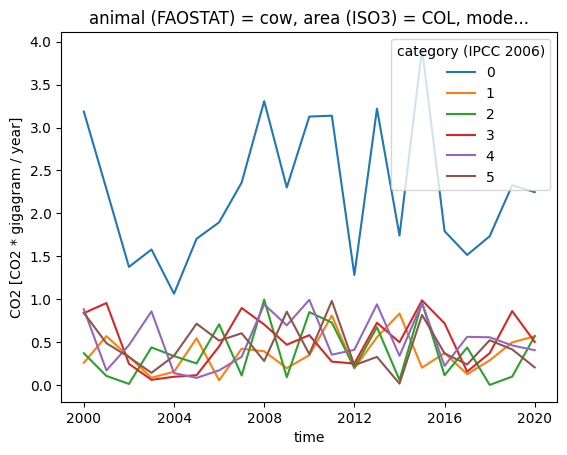
As you can see in the plot, category 0 is now computed from its subcategories. The set() method of Datasets works on all data variables in the dataset which have the corresponding dimension. In this example, the “population” variable does not have categories, so it was unchanged.
Unit handling¶
PRIMAP2 uses the openscm_units package based on the Pint library for handling of units.
CO2 equivalent units and mass units¶
Using global warming potential contexts, it is easy to convert mass units into CO2 equivalents:
[17]:
from primap2 import ureg # The unit registry
sf6_gwp = ds["SF6"].pr.convert_to_gwp(
gwp_context="AR4GWP100", units="Gg CO2 / year"
)
# The information about the used GWP context is retained:
sf6_gwp.attrs
[17]:
{'entity': 'SF6', 'gwp_context': 'AR4GWP100'}
Because the GWP context used for conversion is stored, it is equally easy to convert back to mass units:
[18]:
sf6 = sf6_gwp.pr.convert_to_mass()
The stored GWP context can also be used to convert another array using the same context:
[19]:
ch4_gwp = ds["CH4"].pr.convert_to_gwp_like(sf6_gwp)
Dropping units¶
Sometimes, it is necessary to drop the units, for example to use arrays as input for external functions which are unit-naive. This can be done safely by first converting to the target unit, then dequantifying the dataset or array:
[20]:
ds["CH4"].pint.to("Mt CH4 / year").pr.dequantify()
[20]:
<xarray.DataArray 'CH4' (time: 21, area (ISO3): 4, category (IPCC 2006): 8,
animal (FAOSTAT): 3, product (FAOSTAT): 2,
scenario (FAOSTAT): 2, provenance: 1, model: 1,
source: 2)>
array([[[[[[[[[2.47478594e-03, 2.82506112e-03]]],
[[[1.88801757e-03, 2.47232780e-03]]]],
[[[[3.59865755e-03, 3.53950143e-03]]],
[[[2.80387011e-03, 3.11180055e-03]]]]],
[[[[[3.55444895e-03, 1.93919027e-03]]],
[[[3.44168053e-03, 2.61134035e-03]]]],
...
[[[[4.42050787e-04, 8.05974197e-04]]],
[[[7.29597300e-04, 7.40051386e-04]]]]],
[[[[[1.32828619e-04, 5.59310489e-04]]],
[[[6.14229164e-04, 7.10297284e-04]]]],
[[[[4.55413682e-04, 3.45806115e-04]]],
[[[8.71980975e-04, 5.67013323e-04]]]]]]]]])
Coordinates:
* animal (FAOSTAT) (animal (FAOSTAT)) <U5 'cow' 'swine' 'goat'
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* category (IPCC 2006) (category (IPCC 2006)) object '0' '1' ... '4' '5'
* model (model) <U8 'FANCYFAO'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* provenance (provenance) <U9 'projected'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* source (source) <U8 'RAND2020' 'RAND2021'
* time (time) datetime64[ns] 2000-01-01 ... 2020-01-01
category_names (category (IPCC 2006)) <U14 'total' ... 'land use'
Attributes:
entity: CH4
units: CH4 * megametric_ton / yearNote that the units are then stored in the DataArray’s attrs, and can be restored using the da.pr.quantify function.
Descriptive statistics¶
To get an overview about the missing information in a Dataset or DataArray, you can use the pr.coverage function. It gives you a summary of the number of non-NaN data points:
[21]:
import numpy as np
import xarray as xr
import pandas as pd
time = pd.date_range("2000-01-01", "2003-01-01", freq="AS")
area_iso3 = np.array(["COL", "ARG", "MEX"])
category_ipcc = np.array(["1", "2"])
coords = [
("category (IPCC2006)", category_ipcc),
("area (ISO3)", area_iso3),
("time", time),
]
da = xr.DataArray(
data=[
[
[1, 2, 3, 4],
[np.nan, np.nan, np.nan, np.nan],
[1, 2, 3, np.nan],
],
[
[np.nan, 2, np.nan, 4],
[1, np.nan, 3, np.nan],
[1, np.nan, 3, np.nan],
],
],
coords=coords,
)
da
[21]:
<xarray.DataArray (category (IPCC2006): 2, area (ISO3): 3, time: 4)>
array([[[ 1., 2., 3., 4.],
[nan, nan, nan, nan],
[ 1., 2., 3., nan]],
[[nan, 2., nan, 4.],
[ 1., nan, 3., nan],
[ 1., nan, 3., nan]]])
Coordinates:
* category (IPCC2006) (category (IPCC2006)) <U1 '1' '2'
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX'
* time (time) datetime64[ns] 2000-01-01 ... 2003-01-01[22]:
da.pr.coverage("area")
[22]:
area (ISO3)
COL 6
ARG 2
MEX 5
Name: coverage, dtype: int64
[23]:
da.pr.coverage("time", "area")
[23]:
| area (ISO3) | COL | ARG | MEX |
|---|---|---|---|
| time | |||
| 2000-01-01 | 1 | 1 | 2 |
| 2001-01-01 | 2 | 0 | 1 |
| 2002-01-01 | 1 | 1 | 2 |
| 2003-01-01 | 2 | 0 | 0 |
For Datasets, you can also specify the “entity” as a coordinate to summarize for the data variables:
[24]:
import primap2.tests
ds = primap2.tests.examples.opulent_ds()
ds["CO2"].pr.loc[{"product": "milk"}].pint.magnitude[:] = np.nan
ds.pr.coverage("product", "entity", "area")
[24]:
| area (ISO3) | COL | ARG | MEX | BOL | |
|---|---|---|---|---|---|
| product (FAOSTAT) | entity | ||||
| milk | CO2 | 0 | 0 | 0 | 0 |
| SF6 | 2016 | 2016 | 2016 | 2016 | |
| CH4 | 2016 | 2016 | 2016 | 2016 | |
| SF6 (SARGWP100) | 2016 | 2016 | 2016 | 2016 | |
| meat | CO2 | 2016 | 2016 | 2016 | 2016 |
| SF6 | 2016 | 2016 | 2016 | 2016 | |
| CH4 | 2016 | 2016 | 2016 | 2016 | |
| SF6 (SARGWP100) | 2016 | 2016 | 2016 | 2016 |
Merging¶
xarray provides different functions to combine Datasets and DataArrays. However, these are not built to combine data which contain duplicates with rounding / processing errors. However, when reading data e.g. from country reports this is often needed as some sectors are included in several tables and might use different numbers of decimals. Thus, PRIMAP2 has added a merge function that can accept data discrepancies not exceeding a given tolerance level. The merging of attributes is handled by
xarray and the combine_attrs parameter is just passed on to the xarray functions. Default is to drop_conflicts.
Below an example using the built in opulent_ds
[25]:
from primap2.tests.examples import opulent_ds
import xarray as xr
op_ds = opulent_ds()
# only take part of the countries to have something to actually merge
da_start = op_ds["CO2"].pr.loc[{"area": ["ARG", "COL", "MEX"]}]
# modify some data
data_to_modify = op_ds["CO2"].pr.loc[{"area": ["ARG"]}].pr.sum("area")
data_to_modify.data = data_to_modify.data * 1.009
da_merge = op_ds["CO2"].pr.set(
"area", "ARG", data_to_modify, existing="overwrite"
)
# merge with tolerance such that it will pass
da_result = da_start.pr.merge(da_merge, tolerance=0.01)
da_result
[25]:
<xarray.DataArray 'CO2' (time: 21, area (ISO3): 4, category (IPCC 2006): 8,
animal (FAOSTAT): 3, product (FAOSTAT): 2,
scenario (FAOSTAT): 2, provenance: 1, model: 1,
source: 2)>
<Quantity([[[[[[[[[3.62696857e-01 7.60788785e-01]]]
[[[2.64845489e-02 4.46812952e-01]]]]
[[[[3.71854570e-01 4.77074006e-01]]]
[[[1.27620686e-01 2.22506866e-01]]]]]
[[[[[5.62051590e-01 3.87769116e-01]]]
[[[7.91656206e-01 6.05136589e-01]]]]
...
[[[[3.89570510e-01 7.23844310e-01]]]
[[[6.09056672e-01 6.20044166e-01]]]]]
[[[[[7.23174641e-01 1.32851713e-01]]]
[[[6.61332986e-01 5.02195173e-01]]]]
[[[[4.75262986e-01 8.17348632e-01]]]
[[[2.65430756e-01 2.98518994e-01]]]]]]]]], 'CO2 * gigagram / year')>
Coordinates:
* time (time) datetime64[ns] 2000-01-01 ... 2020-01-01
* area (ISO3) (area (ISO3)) object 'ARG' 'BOL' 'COL' 'MEX'
* category (IPCC 2006) (category (IPCC 2006)) <U3 '0' '1' '2' ... '1.A' '1.B'
* animal (FAOSTAT) (animal (FAOSTAT)) <U5 'cow' 'swine' 'goat'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* provenance (provenance) <U9 'projected'
* model (model) <U8 'FANCYFAO'
* source (source) <U8 'RAND2020' 'RAND2021'
category_names (category (IPCC 2006)) <U14 'total' ... 'light indu...
Attributes:
entity: CO2[26]:
# merge with lower tolerance such that it will fail
try:
da_result = da_start.pr.merge(da_merge, tolerance=0.005)
except xr.MergeError as err:
print(f"An error occured during merging: {err}")
2023-12-12 10:24:50.783 | ERROR | primap2._merge:merge_with_tolerance_core:74 - pr.merge error: found discrepancies larger than tolerance (0.50%) for area (ISO3)=ARG, provenance=projected, model=FANCYFAO:
shown are relative discrepancies.
category_names CO2
time category (IPCC 2006) animal (FAOSTAT) product (FAOSTAT) scenario (FAOSTAT) source
2000-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2001-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2002-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2003-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2004-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2005-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2006-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2007-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2008-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2009-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2010-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2011-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2012-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2013-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2014-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2015-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2016-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2017-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2018-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2019-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2020-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
An error occured during merging: pr.merge error: found discrepancies larger than tolerance (0.50%) for area (ISO3)=ARG, provenance=projected, model=FANCYFAO:
shown are relative discrepancies.
category_names CO2
time category (IPCC 2006) animal (FAOSTAT) product (FAOSTAT) scenario (FAOSTAT) source
2000-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2001-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2002-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2003-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2004-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2005-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2006-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2007-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2008-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2009-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2010-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2011-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2012-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2013-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2014-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2015-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2016-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2017-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2018-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2019-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2020-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
[27]:
# only throw a warning and not an error
da_result = da_start.pr.merge(
da_merge, tolerance=0.005, error_on_discrepancy=False
)
da_result
2023-12-12 10:24:52.995 | WARNING | primap2._merge:merge_with_tolerance_core:78 - pr.merge error: found discrepancies larger than tolerance (0.50%) for area (ISO3)=ARG, provenance=projected, model=FANCYFAO:
shown are relative discrepancies.
category_names CO2
time category (IPCC 2006) animal (FAOSTAT) product (FAOSTAT) scenario (FAOSTAT) source
2000-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2001-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2002-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2003-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2004-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2005-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2006-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2007-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2008-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2009-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2010-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2011-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2012-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2013-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2014-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2015-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2016-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2017-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2018-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2019-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
2020-01-01 0 cow milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
swine milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
goat milk highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
meat highpop RAND2020 total 0.009
RAND2021 total 0.009
lowpop RAND2020 total 0.009
RAND2021 total 0.009
1 cow milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
swine milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
goat milk highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
meat highpop RAND2020 industry 0.009
RAND2021 industry 0.009
lowpop RAND2020 industry 0.009
RAND2021 industry 0.009
2 cow milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
swine milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
goat milk highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
meat highpop RAND2020 energy 0.009
RAND2021 energy 0.009
lowpop RAND2020 energy 0.009
RAND2021 energy 0.009
3 cow milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
swine milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
goat milk highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
meat highpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
lowpop RAND2020 transportation 0.009
RAND2021 transportation 0.009
4 cow milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
swine milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
goat milk highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
meat highpop RAND2020 residential 0.009
RAND2021 residential 0.009
lowpop RAND2020 residential 0.009
RAND2021 residential 0.009
5 cow milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
swine milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
goat milk highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
meat highpop RAND2020 land use 0.009
RAND2021 land use 0.009
lowpop RAND2020 land use 0.009
RAND2021 land use 0.009
1.A cow milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
swine milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
goat milk highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
meat highpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
lowpop RAND2020 heavy industry 0.009
RAND2021 heavy industry 0.009
1.B cow milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
swine milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
goat milk highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
meat highpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
lowpop RAND2020 light industry 0.009
RAND2021 light industry 0.009
[27]:
<xarray.DataArray 'CO2' (time: 21, area (ISO3): 4, category (IPCC 2006): 8,
animal (FAOSTAT): 3, product (FAOSTAT): 2,
scenario (FAOSTAT): 2, provenance: 1, model: 1,
source: 2)>
<Quantity([[[[[[[[[3.62696857e-01 7.60788785e-01]]]
[[[2.64845489e-02 4.46812952e-01]]]]
[[[[3.71854570e-01 4.77074006e-01]]]
[[[1.27620686e-01 2.22506866e-01]]]]]
[[[[[5.62051590e-01 3.87769116e-01]]]
[[[7.91656206e-01 6.05136589e-01]]]]
...
[[[[3.89570510e-01 7.23844310e-01]]]
[[[6.09056672e-01 6.20044166e-01]]]]]
[[[[[7.23174641e-01 1.32851713e-01]]]
[[[6.61332986e-01 5.02195173e-01]]]]
[[[[4.75262986e-01 8.17348632e-01]]]
[[[2.65430756e-01 2.98518994e-01]]]]]]]]], 'CO2 * gigagram / year')>
Coordinates:
* time (time) datetime64[ns] 2000-01-01 ... 2020-01-01
* area (ISO3) (area (ISO3)) object 'ARG' 'BOL' 'COL' 'MEX'
* category (IPCC 2006) (category (IPCC 2006)) <U3 '0' '1' '2' ... '1.A' '1.B'
* animal (FAOSTAT) (animal (FAOSTAT)) <U5 'cow' 'swine' 'goat'
* product (FAOSTAT) (product (FAOSTAT)) <U4 'milk' 'meat'
* scenario (FAOSTAT) (scenario (FAOSTAT)) <U7 'highpop' 'lowpop'
* provenance (provenance) <U9 'projected'
* model (model) <U8 'FANCYFAO'
* source (source) <U8 'RAND2020' 'RAND2021'
category_names (category (IPCC 2006)) <U14 'total' ... 'light indu...
Attributes:
entity: CO2Aggregation and infilling¶
xarray provides robust functions for aggregation (sum) and filling of missing information (fillna). PRIMAP2 adds functions which fill or skip missing information based on if the information is missing at all points along certain axes, for example for a whole time series. This makes it possible to, for example, evaluate the sum of sub-categories while ignoring only those categories which are missing completely. It is also possible to ignore NA values (i.e. treating them as 0) in sums
using the skipna parameter. When using skipna, the min_count parameter governs how many non-NA vales are needed in a sum for the result to be non-NA. The default value is skipna=1. This is helpful if you want to e.g. sum all subsectors and for some countries or gases some of the subsectors have NA values because there is no data. To avoid NA timeseries if a single sector is NA you use skipna. In other cases, e.g. when checking if data coverage is complete skipna is not
used, so any NA value in the source data results in NA in the summed data and is not hidden.
[28]:
time = pd.date_range("2000-01-01", "2003-01-01", freq="AS")
area_iso3 = np.array(["COL", "ARG", "MEX"])
coords = [("area (ISO3)", area_iso3), ("time", time)]
da = xr.DataArray(
data=[
[1, 2, 3, 4],
[np.nan, np.nan, np.nan, np.nan],
[1, 2, 3, np.nan],
],
coords=coords,
)
da
[28]:
<xarray.DataArray (area (ISO3): 3, time: 4)>
array([[ 1., 2., 3., 4.],
[nan, nan, nan, nan],
[ 1., 2., 3., nan]])
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2003-01-01[29]:
da.pr.sum(dim="area", skipna_evaluation_dims="time")
[29]:
<xarray.DataArray (time: 4)> array([ 2., 4., 6., nan]) Coordinates: * time (time) datetime64[ns] 2000-01-01 2001-01-01 2002-01-01 2003-01-01
[30]:
da.pr.sum(dim="area", skipna=True)
[30]:
<xarray.DataArray (time: 4)> array([2., 4., 6., 4.]) Coordinates: * time (time) datetime64[ns] 2000-01-01 2001-01-01 2002-01-01 2003-01-01
[31]:
# compare this to the result of the standard xarray sum:
da.sum(dim="area (ISO3)")
[31]:
<xarray.DataArray (time: 4)> array([2., 4., 6., 4.]) Coordinates: * time (time) datetime64[ns] 2000-01-01 2001-01-01 2002-01-01 2003-01-01
The same functionality is available for filling in missing information:
[32]:
da.pr.fill_all_na("time", value=10)
[32]:
<xarray.DataArray (area (ISO3): 3, time: 4)>
array([[ 1., 2., 3., 4.],
[10., 10., 10., 10.],
[ 1., 2., 3., nan]])
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2003-01-01Downscaling¶
To downscale a super-category (for example, regional data) to sub-categories (for example, country-level data in the same region), the downscale_timeseries function is available:
[33]:
# select an example dataset
da = primap2.open_dataset("minimal_ds.nc")["CO2"].loc[
{"time": slice("2000", "2010")}
]
da
[33]:
<xarray.DataArray 'CO2' (time: 11, area (ISO3): 4, source: 1)>
<Quantity([[[0.66352018]
[0.91683183]
[0.10638745]
[0.61416341]]
[[0.33822533]
[0.68233863]
[0.73953851]
[0.79057726]]
[[0.11409101]
[0.25072807]
[0.85044897]
[0.40873907]]
[[0.80147707]
[0.85209597]
[0.71521289]
[0.08031265]]
...
[[0.65464417]
[0.26581567]
[0.96899088]
[0.43650759]]
[[0.6612736 ]
[0.59752662]
[0.78509942]
[0.55512166]]
[[0.33259991]
[0.05037719]
[0.33692108]
[0.65173881]]
[[0.55220105]
[0.91061979]
[0.82348029]
[0.52753096]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
entity: CO2[34]:
# compute regional data as sum of country-level data
temp = da.sum(dim="area (ISO3)")
temp = temp.expand_dims({"area (ISO3)": ["LATAM"]})
# delete data from the country level for the years 2005-2009 (inclusive)
da.loc[{"time": slice("2005", "2009")}].pint.magnitude[:] = np.nan
# add regional data to the array
da = xr.concat([da, temp], dim="area (ISO3)")
da
[34]:
<xarray.DataArray 'CO2' (time: 11, area (ISO3): 5, source: 1)>
<Quantity([[[0.66352018]
[0.91683183]
[0.10638745]
[0.61416341]
[2.30090286]]
[[0.33822533]
[0.68233863]
[0.73953851]
[0.79057726]
[2.55067973]]
[[0.11409101]
[0.25072807]
[0.85044897]
[0.40873907]
[1.62400712]]
[[0.80147707]
[0.85209597]
...
[ nan]
[2.32595831]]
[[ nan]
[ nan]
[ nan]
[ nan]
[2.5990213 ]]
[[ nan]
[ nan]
[ nan]
[ nan]
[1.37163699]]
[[0.55220105]
[0.91061979]
[0.82348029]
[0.52753096]
[2.81383208]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) object 'COL' 'ARG' 'MEX' 'BOL' 'LATAM'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
entity: CO2[35]:
# Do the downscaling
da.pr.downscale_timeseries(
basket="LATAM",
basket_contents=["BOL", "MEX", "COL", "ARG"],
dim="area (ISO3)",
)
[35]:
<xarray.DataArray 'CO2' (time: 11, area (ISO3): 5, source: 1)>
<Quantity([[[0.66352018]
[0.91683183]
[0.10638745]
[0.61416341]
[2.30090286]]
[[0.33822533]
[0.68233863]
[0.73953851]
[0.79057726]
[2.55067973]]
[[0.11409101]
[0.25072807]
[0.85044897]
[0.40873907]
[1.62400712]]
[[0.80147707]
[0.85209597]
...
[0.27789765]
[2.32595831]]
[[0.5851001 ]
[0.91460464]
[0.72993588]
[0.36938068]
[2.5990213 ]]
[[0.28895518]
[0.46326178]
[0.39333118]
[0.22608885]
[1.37163699]]
[[0.55220105]
[0.91061979]
[0.82348029]
[0.52753096]
[2.81383208]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) object 'COL' 'ARG' 'MEX' 'BOL' 'LATAM'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
entity: CO2For the downscaling, shares for the sub-categories at the points in time where data for all sub-categories is available are determined, the shares are interpolated where data is missing, and then the super-category is downscaled using these shares.
Handling of gas baskets¶
Summation¶
To sum the contents of gas baskets like KYOTOGHG, the function ds.gas_basket_contents_sum is available:
[36]:
# select example dataset
ds = primap2.open_dataset("minimal_ds.nc").loc[
{"time": slice("2000", "2010")}
]
ds
[36]:
<xarray.Dataset>
Dimensions: (time: 11, area (ISO3): 4, source: 1)
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Data variables:
CH4 (time, area (ISO3), source) float64 [CH4·Gg/a] 0.7543 .....
CO2 (time, area (ISO3), source) float64 [CO2·Gg/a] 0.6635 .....
SF6 (time, area (ISO3), source) float64 [Gg·SF6/a] 0.0018 .....
SF6 (SARGWP100) (time, area (ISO3), source) float64 [CO2·Gg/a] 43.02 ......
Attributes:
area: area (ISO3)[37]:
# add (empty) gas basket with corresponding metadata
ds["KYOTOGHG"] = xr.full_like(ds["CO2"], np.nan).pr.quantify(
units="Gg CO2 / year"
)
ds["KYOTOGHG"].attrs = {"entity": "KYOTOGHG", "gwp_context": "AR4GWP100"}
ds
[37]:
<xarray.Dataset>
Dimensions: (time: 11, area (ISO3): 4, source: 1)
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Data variables:
CH4 (time, area (ISO3), source) float64 [CH4·Gg/a] 0.7543 .....
CO2 (time, area (ISO3), source) float64 [CO2·Gg/a] 0.6635 .....
SF6 (time, area (ISO3), source) float64 [Gg·SF6/a] 0.0018 .....
SF6 (SARGWP100) (time, area (ISO3), source) float64 [CO2·Gg/a] 43.02 ......
KYOTOGHG (time, area (ISO3), source) float64 [CO2·Gg/a] nan ... nan
Attributes:
area: area (ISO3)[38]:
# compute gas basket from its contents, which have to be given explicitly
ds.pr.gas_basket_contents_sum(
basket="KYOTOGHG",
basket_contents=["CO2", "SF6", "CH4"],
)
[38]:
<xarray.DataArray 'KYOTOGHG' (time: 11, area (ISO3): 4, source: 1)>
<Quantity([[[ 60.55826022]
[11537.15564864]
[15441.09112246]
[ 2832.13708223]]
[[10913.89923578]
[12017.91242534]
[18293.41941003]
[18461.14522004]]
[[12164.44190136]
[14106.2809531 ]
[ 5472.05066779]
[ 7054.12526007]]
[[12163.56107579]
[11353.23339466]
[ 9916.31088755]
[15594.44908077]]
...
[[ 2626.6977983 ]
[14671.83782755]
[14756.40772611]
[18365.25904294]]
[[ 3531.11691325]
[11891.14993374]
[ 2029.28907109]
[14178.86679903]]
[[20526.74343151]
[ 3892.41005405]
[14325.27379526]
[ 8953.63696907]]
[[22693.36709336]
[ 5369.92103899]
[21612.22026121]
[17281.9212453 ]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
gwp_context: AR4GWP100
entity: KYOTOGHGNote that like all PRIMAP2 functions, gas_basket_contents_sum returns the result without overwriting anything in the original dataset, so you have to explicitly overwrite existing data if you want that:
[39]:
ds["KYOTOGHG"] = ds.pr.gas_basket_contents_sum(
basket="KYOTOGHG",
basket_contents=["CO2", "SF6", "CH4"],
)
Filling in missing information¶
To fill in missing data in a gas basket, use fill_na_gas_basket_from_contents:
[40]:
# delete all data about the years 2005-2009 (inclusive) from the
# KYOTOGHG data
ds["KYOTOGHG"].loc[{"time": slice("2005", "2009")}].pint.magnitude[
:
] = np.nan
ds["KYOTOGHG"]
[40]:
<xarray.DataArray 'KYOTOGHG' (time: 11, area (ISO3): 4, source: 1)>
<Quantity([[[ 60.55826022]
[11537.15564864]
[15441.09112246]
[ 2832.13708223]]
[[10913.89923578]
[12017.91242534]
[18293.41941003]
[18461.14522004]]
[[12164.44190136]
[14106.2809531 ]
[ 5472.05066779]
[ 7054.12526007]]
[[12163.56107579]
[11353.23339466]
[ 9916.31088755]
[15594.44908077]]
...
[[ nan]
[ nan]
[ nan]
[ nan]]
[[ nan]
[ nan]
[ nan]
[ nan]]
[[ nan]
[ nan]
[ nan]
[ nan]]
[[22693.36709336]
[ 5369.92103899]
[21612.22026121]
[17281.9212453 ]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
gwp_context: AR4GWP100
entity: KYOTOGHG[41]:
ds.pr.fill_na_gas_basket_from_contents(
basket="KYOTOGHG", basket_contents=["CO2", "SF6", "CH4"]
)
[41]:
<xarray.DataArray 'KYOTOGHG' (time: 11, area (ISO3): 4, source: 1)>
<Quantity([[[ 60.55826022]
[11537.15564864]
[15441.09112246]
[ 2832.13708223]]
[[10913.89923578]
[12017.91242534]
[18293.41941003]
[18461.14522004]]
[[12164.44190136]
[14106.2809531 ]
[ 5472.05066779]
[ 7054.12526007]]
[[12163.56107579]
[11353.23339466]
[ 9916.31088755]
[15594.44908077]]
...
[[ 2626.6977983 ]
[14671.83782755]
[14756.40772611]
[18365.25904294]]
[[ 3531.11691325]
[11891.14993374]
[ 2029.28907109]
[14178.86679903]]
[[20526.74343151]
[ 3892.41005405]
[14325.27379526]
[ 8953.63696907]]
[[22693.36709336]
[ 5369.92103899]
[21612.22026121]
[17281.9212453 ]]], 'CO2 * gigagram / year')>
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Attributes:
gwp_context: AR4GWP100
entity: KYOTOGHGThe reverse case is that you are missing some data in the timeseries of individual gases and want to fill those in using downscaled data from a gas basket. Here, use downscale_gas_timeseries:
[42]:
# delete all data about the years 2005-2009 from the individual gas data
sel = {"time": slice("2005", "2009")}
ds["CO2"].loc[sel].pint.magnitude[:] = np.nan
ds["SF6"].loc[sel].pint.magnitude[:] = np.nan
ds["CH4"].loc[sel].pint.magnitude[:] = np.nan
ds
[42]:
<xarray.Dataset>
Dimensions: (time: 11, area (ISO3): 4, source: 1)
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Data variables:
CH4 (time, area (ISO3), source) float64 [CH4·Gg/a] 0.7543 .....
CO2 (time, area (ISO3), source) float64 [CO2·Gg/a] 0.6635 .....
SF6 (time, area (ISO3), source) float64 [Gg·SF6/a] 0.0018 .....
SF6 (SARGWP100) (time, area (ISO3), source) float64 [CO2·Gg/a] 43.02 ......
KYOTOGHG (time, area (ISO3), source) float64 [CO2·Gg/a] 60.56 ......
Attributes:
area: area (ISO3)[43]:
# This determines gas shares at the points in time where individual gas
# data is available, interpolates the shares where data is missing, and
# then downscales the gas basket data using the interpolated shares
ds.pr.downscale_gas_timeseries(
basket="KYOTOGHG", basket_contents=["CO2", "SF6", "CH4"]
)
[43]:
<xarray.Dataset>
Dimensions: (time: 11, area (ISO3): 4, source: 1)
Coordinates:
* area (ISO3) (area (ISO3)) <U3 'COL' 'ARG' 'MEX' 'BOL'
* source (source) <U8 'RAND2020'
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2010-01-01
Data variables:
CH4 (time, area (ISO3), source) float64 [CH4·Gg/a] 0.7543 .....
CO2 (time, area (ISO3), source) float64 [CO2·Gg/a] 0.6635 .....
SF6 (time, area (ISO3), source) float64 [Gg·SF6/a] 0.0018 .....
SF6 (SARGWP100) (time, area (ISO3), source) float64 [CO2·Gg/a] 43.02 ......
KYOTOGHG (time, area (ISO3), source) float64 [CO2·Gg/a] 60.56 ......
Attributes:
area: area (ISO3)